Mining Drug-Drug Interaction Induced Adverse Effects from Health Record Databases
National Science Foundation Project
Award Number: 1622526, 1827472
Investigators: Li Shen, Xia Ning, and Lang Li
NSF Progarm: Smart and Connected Health
Abstract
Recent advances in large-scale electronic health record database techniques provide exciting new opportunities to the study of drug safety. Drug-drug interactions (DDIs), a major cause of adverse drug events (ADEs), are a serious global health concern, and a severe detriment to public health. The scale of DDIs involving three or more drugs (also called high-order DDIs) has posed a prohibitory challenge for its molecular pharmacology and clinical research, which motivates alternative strategies such as mining health record data. This project aims to develop large-scale computational strategies and effective software tools for mining high-order DDI effects from health record databases, in order to yield novel discoveries in drug safety, and ultimately to benefit national health and well being.
To achieve the above goal, this project is designed to complete four specific tasks. Task 1 aims to develop a novel statistical framework to discover high-order DDI signals associated with ADEs from health record databases. Task 2 aims to study a novel drug safety problem for mining directional DDI signals. Task 3 aims to develop an innovative approach for mining directional DDI patterns at the drug-group level. Task 4 is devoted to software development, evaluation and validation. The project applies these methods to analyze three independent databases, packages method implementations into a user-friendly software toolkit, and releases the toolkit to the public. This project not only facilitates the development of novel computational techniques in drug safety research, but also addresses emerging scientific questions in modeling, mining, and visual exploration of complex data such as the health record data. The project's educational activities include course development, student mentoring and advising, and involvement of minority and underrepresented students in research activities.
Products
Junfeng Liu and Xia Ning "Differential Compound Prioritization via Bi-Directional Selectivity Push with Power" Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics , 2017 , p.394-399 10.1145/3107411.3107486
Junfeng Liu and Xia Ning "Multi-Assay-Based Compound Prioritization via Assistance Utilization: A Machine Learning Framework" Chemical Information and Modeling , 2017 , p.484-498 10.1021/acs.jcim.6b00737
Ning X, Li L, Shen L. (2017) Pattern discovery from directional high-order drug-drug interaction relations. NetSci’17: Int. School and Conf. on Network Science, Indianapolis, IN, June 21-23, 2017.
Ning X, Schleyer T, Shen L, Li L. (2017) Pattern discovery from directional high-order drug-drug interaction relations. ICHI’17: The 5th IEEE International Conference on Healthcare Informatics, 9 pages, Park City, Utah, August 23-26, 2017
Ning X, Shen L, Li L. (2017) Predicting high-order directional drug-drug interaction relations. ICHI’17: The 5th IEEE International Conference on Healthcare Informatics, 6 pages, Park City, Utah, August 23-26, 2017.
Chiang C, Zhang P, Wang X, Wang L, Zhang S, Ning X, Shen L, Quinney S, Li L. (2018) Translational high-dimensional drug Interaction discovery and validation using health record databases and pharmacokinetics models. Clinical Pharmacology & Therapeutics, 103(2):287-95.
Chiang WH, Schleyer T, Shen L, Li L, Ning X. (2018) Pattern discovery from high-order drug-drug interaction relations. Journal of Healthcare Informatics Research, 2(3):272-304.
Liu J, Ning X "Differential compound prioritization via bi-directional selectivity push with power" Journal of Chemical Information and Modeling , v.57 , 2017 , p.2958
Wang X, Zhang P, Chiang C, Wu H, Shen L, Ning X, Zeng D, Wang L, Quinney SK, Feng W, Li L. (2018) Mixture drug-count response model for the high dimensional drug combinatory effect on myopathy. Statistics in Medicine, 37(4):673-86.
Chasioti D, Yao X, Zhang P, Ning X, Li L, Shen L. (2017) Mining directional drug interaction effects on myopathy using the FAERS database. PSB’17: Pac Symp Biocomput., poster #71, Big Island of Hawaii, January 3-7, 2017.
Chasioti D, Yao X, Zhang P, Quinney S, Ning X, Li L, Shen L. (2017) Mining and visualizing the network of directional drug interaction effects. NetSci’17: Int. School and Conf. on Network Science, Indianapolis, IN, June 21-23, 2017.
He Y, Liu J, Ning X. (2018) Drug selection via joint push and learning to rank. IEEE Transactions on Computational Biology and Bioinformatics., in press. DOI: 10.1109/TCBB.2018.2848908
Zhang P, Li M, Chiang CW, Wang L, Xiang Y, Cheng L, Feng W, Schleyer TK, Quinney SK, Wu HY, Zeng D, Li L. (2018) Three-Component Mixture Model-Based Adverse Drug Event Signal Detection for the Adverse Event Reporting System. CPT Pharmacometrics Syst Pharmacol., 7(8), 499-506. doi: 10.1002/psp4.12294
Chiang W, Shen L, Li L, Ning X. (2018) Drug-drug interaction prediction based on co-medication patterns and graph matching. International Journal of Computational Biology and Drug Design, 13(1):36 – 57.
Yao X, Tsang T, Quinney S, Zhang P, Ning X, Li L, Shen L. (2019) Mining and visualizing high-order directional drug interaction effects using the FAERS database. ICIBM’19: Int. Conf. on Intelligent Biology and Medicine, Columbus, OH, USA, June 9-11, 2019.
Yao X, Tsang T, Quinney S, Zhang P, Ning X, Li L, Shen L. (2020) Mining and visualizing high-order directional drug interaction effects using the FAERS database. BMC Medical Informatics and Decision Making, 20(Suppl 2):50.
Peng B, Ning X. (2019) Deep learning for high-order drug-drug interaction prediction. BCB '19: Proceedings of the 10th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, September 2019 Pages 197–206. DOI: 10.1145/3307339.3342136
Chasioti D, Yao X, Zhang P, Lerner S, Quinney SK, Ning X, Li L, Shen L. (2019) Mining directional drug interaction effects on myopathy using the FAERS database. IEEE J Biomed Health Inform, 23(5):2156-2163. doi: 10.1109/JBHI.2018.2874533
Zhu A, Zeng D, Shen L, Ning X, Li L, Zhang P "A Super-combo-drug test (SupCD-T) to Detect Adverse Drug Events and Drug Interactions from Electronic Health Records in the Era of Polypharmacy" Statistics in Medicine , v.39 , 2020 , p.1458 10.1002/sim.8490
Wang L, Shendre A, Chiang CW, Cao W, Ning X, Zhang P, Zhang P, Li L "A pharmacovigilance study of pharmacokinetic drug interactions using a translational informatics discovery approach" Br J Clin Pharmacol , 2021 doi: 10.1111/bcp.14762
Xia Ning , Ziwei Fan, Evan Burgun, Zhiyun Ren, and Titus Schleyer "Improving information retrieval from electronic health records using dynamic and multi-collaborative filtering" Plos One , v.16 , 2021 , p.e0255467 10.1371/journal.pone.0255467
Zhiyun Ren, Bo Peng, Titus Schleyer, and Xia Ning "Hybrid collaborative filtering methods for recommending search terms to clinicians" Journal of Biomedical Informatics , v.113 , 2020 , p.103635 10.1016/j.jbi.2020.103635
Ziqi Chen, Martin Renqiang Min, and Xia Ning "Ranking-based convolutional neural network models for peptide-MHC Class I binding prediction" Frontiers in Molecular Biosciences , v.8 , 2021 , p.634836 10.3389/fmolb.2021.634836
Dey, Vishal and Machiraju, Raghu and Ning, Xia (2022). Improving Compound Activity Classification via Deep Transfer and Representation Learning. ACS Omega. 7 (11), 9465-9483. DOI: 10.1021/acsomega.1c06805
Peng, Bo and Ren, Zhiyun and Parthasarathy, Srinivasan and Ning, Xia (2022). HAM: Hybrid Associations Models for Sequential Recommendation. IEEE Transactions on Knowledge and Data Engineering. 34 (10), 4838-4853. DOI: 10.1109/TKDE.2021.3049692
Peng, Bo and Ren, Zhiyun and Parthasarathy, Srinivasan and Ning, Xia (2022). M2: Mixed Models with Preferences, Popularities and Transitions for Next-Basket Recommendation. IEEE Transactions on Knowledge and Data Engineering. 1-1. DOI: 10.1109/TKDE.2022.3142773
Ziqi Chen, Martin Renqiang Min, Srinivasan Parthasarathy, and Xia Ning (2021). A deep generative model for molecule optimization via one fragment modification. Nature Machine Intelligence. 3 1040–1049. DOI: https://doi.org/10.1038/s42256-021-00410-2
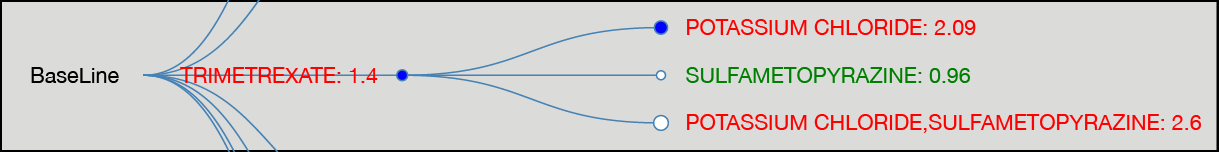
DDI Visualization: Test our DDI Explorer.
D3I Code: Code and data for "Deep Learning for High-Order Drug-Drug Interaction Prediction"