Spherical Parameterization¶
Spherical parameterization creates a continuous and uniform mapping from the object surface to the surface of a unit sphere, and its result is a bijective mapping between each point 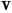 on a surface and a pair of spherical coordinates
on a surface and a pair of spherical coordinates 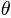 and
and 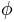 :
:
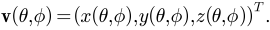
The spherical parameterization method proposed with the original SPHARM paper [Brechb1995] aimed to create an equal area mapping as well as minimize angle distortions by solving a constrained optimization problem, which was implemented in SPHARM-PDM [Styner2006]. This traditional method has been very effective in analyzing small and moderately-sized structures extracted from volumetric images (e.g., MRI, CT). However, it is applicable only to voxel surfaces. CALD [Shen2006] is a newer method that extends the traditional method and can be applied to general triangular meshes. In SPHARM-MAT, an enhanced version of CALD method is available for analyzing triangular meshes (see Exercise 3.1 Surface Meshes (CALD) and Exercise 3.2 Voxel Surfaces (CALD)). In addition, SPHARM-PDM can also be called from SPHARM-MAT for analyzing voxel surfaces (see Exercise 3.3 Voxel Surfaces (PDM)).
Area Distortion Cost¶
The concept of area distortion cost introduced by [Shen2006] is employed as the performance measure in the following exercises. Let 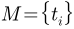 be a surface mesh in the object space and let
be a surface mesh in the object space and let  be its spherical parameterization, which maps
be its spherical parameterization, which maps 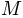 to the unit sphere
to the unit sphere 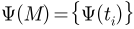 .
.
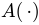 is used to denote the relative area of a triangle or a spherical triangle:
is used to denote the relative area of a triangle or a spherical triangle:
- For a triangle, its relative area is defined as its area divided by the total area of the object surface.
- For a spherical triangle, its relative area is defined as its area divided by
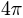 , the surface area of the unit sphere.
, the surface area of the unit sphere.
The area distortion cost (ADC) 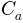 with respect to
with respect to 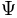 is defined as follows:
is defined as follows:
For each triangle
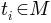 ,
,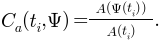
This measures the local ADC of a single triangle.
For the whole parametric mesh
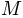 ,
,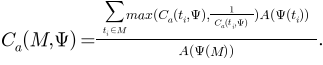
This measures the overall ADC for the whole mesh. By taking
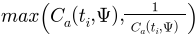
as the ADC contribution from each triangle, we treat contraction and expansion equally, and so always have
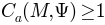 .
.
Exercise 3.1 Surface Meshes (CALD)¶
This exercise was tested on a WinXP machine (3GHz CPU, 3.25G RAM) running Matlab 7.7.0 (R2008b). It took a few minutes to finish.
Major Steps
- CALD Parameterization for Surface Meshes
- Surface Visualization (ADC_ParaMap, Solid with Mesh)
CALD Parameterization for Surface Meshes¶
Task
Create spherical parameterization for surface meshes
Input
SpharmMatDir/data/Ex0301/mesh01_obj/*_obj.mat: An input surface needs to be genus zero (i.e., with spherical topology), and can be represented by a triangulation or a quadrilateral mesh.
Output
SpharmMatDir/data/Ex0301/mesh02_smo/initParamCALD/*_ini.mat: Results of initial parameterization
SpharmMatDir/data/Ex0301/mesh02_smo/*_smo.mat: Results of optimized parameterization
Steps
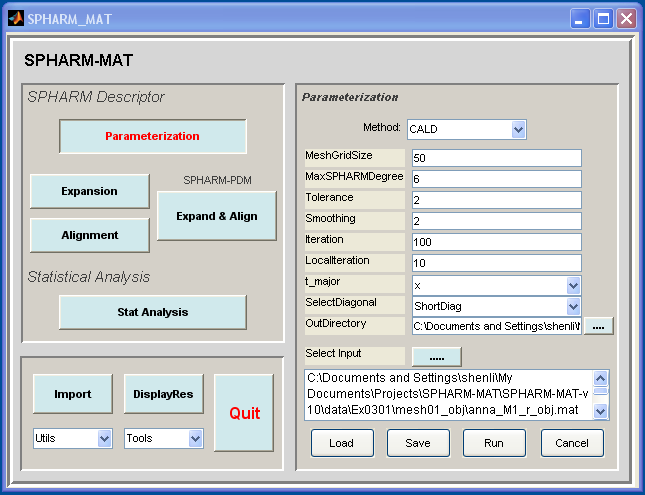
- Make an output directory SpharmMatDir/data/Ex0301/mesh02_smo
- Run SPHARM_MAT.m under Matlab
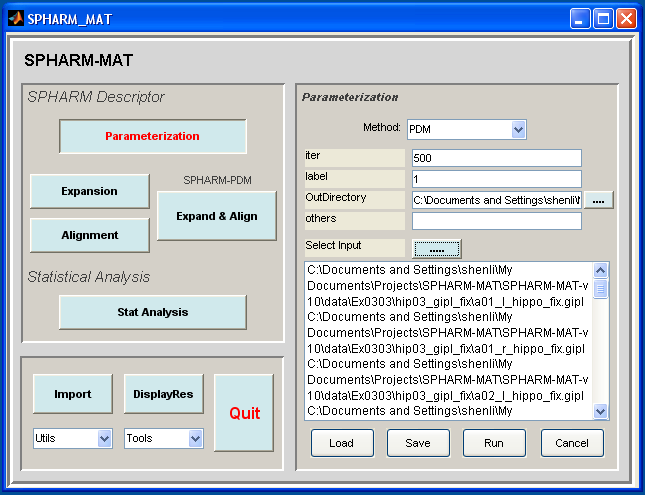
- Click Parameterization button
- In the right panel, let Method be CALD
- In the right panel, let MeshGridSize be 50, MaxSPHARMDegree be 6, Tolerance be 2, Smoothing be 2, Iteration be 100, LocalIteration be 10, t_major be x, SelectDiagonal be ShortDiag.
- In the right panel, select SpharmMatDir/data/Ex0301/mesh02_smo as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *_obj.mat files under SpharmMatDir/data/Ex0301/mesh01_obj as input files
- Click OK button (See Screen Capture for CALD Parameterization (Surface Mesh))
Surface Visualization (ADC_ParaMap, Solid with Mesh)¶
Task
Visualize surface meshes in both object and parameter spaces
Input
SpharmMatDir/data/Ex0301/mesh02_smo/*_smo.mat or
SpharmMatDir/data/Ex0301/mesh02_smo/initParamCALD/*_ini.mat
Output
SpharmMatDir/data/Ex0301/mesh02_smo/PNG/*.png or
SpharmMatDir/data/Ex0301/mesh02_smo/initParamCALD/PNG/*.png
Steps
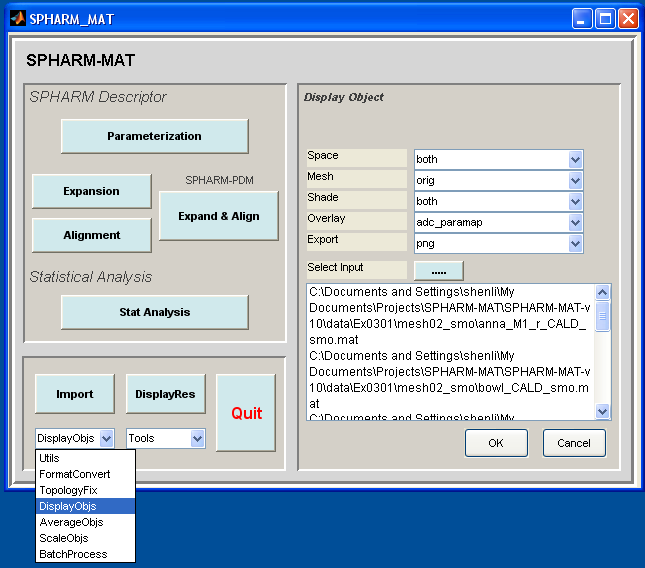
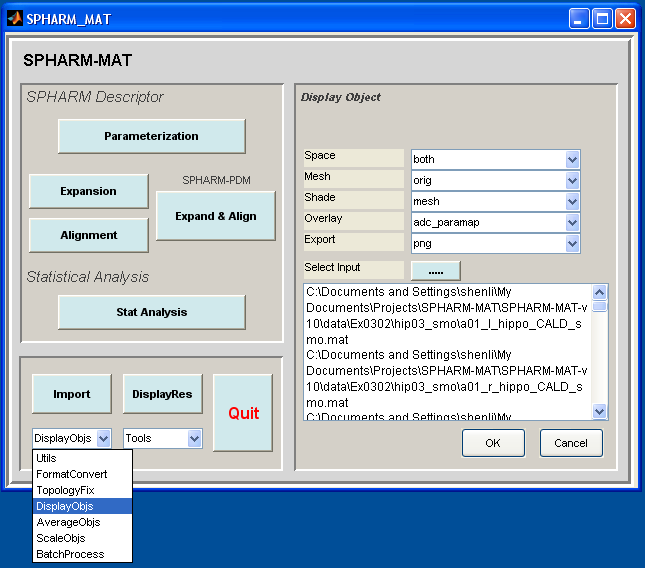
- Select DisplayObjs under the Utils pop-up menu (bottom-left corner)
- In the right panel, let Space be both, Mesh be orig, Shade be both, Overlay be adc_paramap, Export be PNG
- In the right panel, click ..... button next to Select Input, and select either all the SpharmMatDir/data/Ex0301/mesh02_smo/PNG/*.png files or all the SpharmMatDir/data/Ex0301/mesh02_smo/initParamCALD/PNG/*.png as the input files
- Click OK button (See Screen Capture for Surface Visualization (ADC_ParaMap, Solid with Mesh))
Notes
- The visualization results are saved as PNG files under SpharmMatDir/data/Ex0301/mesh02_smo/PNG and SpharmMatDir/data/Ex0301/mesh02_smo/initParamCALD/PNG.
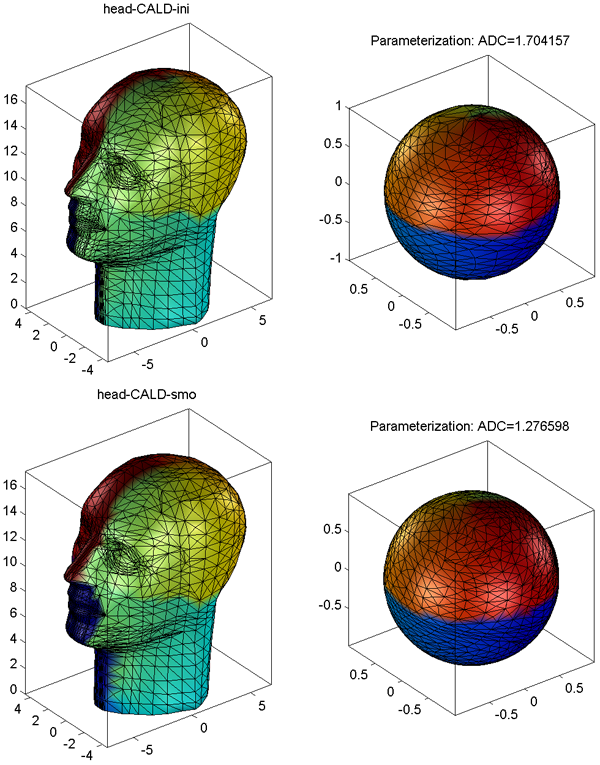
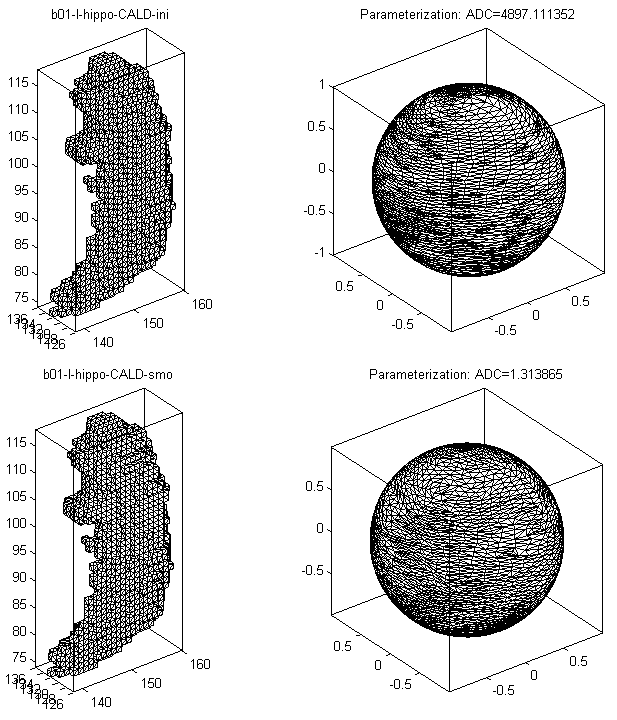
- The results of initial parameterization and optimized parameterization for the head model are shown in Surface Visualization (ADC_ParaMap, Solid with Mesh), where the ADC (area distortion cost) is defined in Area Distortion Cost.
Exercise 3.2 Voxel Surfaces (CALD)¶
This exercise was tested on a WinXP machine (3GHz CPU, 3.25G RAM) running Matlab 7.7.0 (R2008b). It took 10-20 minutes to finish.
Major Steps
- CALD Parameterization for Voxel Surfaces
- Surface Visualization (ADC_ParaMap, Mesh)
CALD Parameterization for Voxel Surfaces¶
Task
Create spherical parameterization for voxel surfaces using CALD
Input
SpharmMatDir/data/Ex0302/hip02_fix/*_fix.mat: This folder is a copy of SpharmMatDir/data/Ex0201/hip02_fix/*_fix.mat. The surface of an input binary object needs to be genus zero (i.e., with spherical topology).
Output
SpharmMatDir/data/Ex0302/hip03_smo/initParamCALD/*_ini.mat: Results of initial parameterization
SpharmMatDir/data/Ex0302/hip03_smo/*_smo.mat: Results of optimized parameterization
Steps
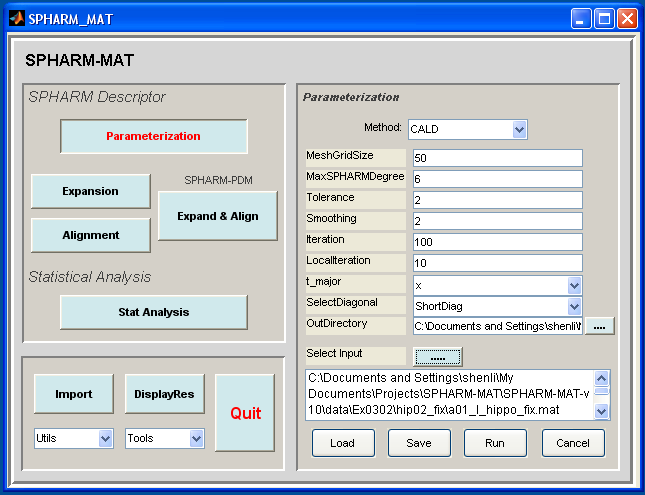
- Make an output directory SpharmMatDir/data/Ex0302/hip03_smo
- Run SPHARM_MAT.m under Matlab
- Click Parameterization button
- In the right panel, let Method be CALD
- In the right panel, let MeshGridSize be 50, MaxSPHARMDegree be 6, Tolerance be 2, Smoothing be 2, Iteration be 100, LocalIteration be 10, t_major be x, SelectDiagonal be ShortDiag.
- In the right panel, select SpharmMatDir/data/Ex0302/hip03_smo as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *_fix.mat files under SpharmMatDir/data/Ex0302/hip02_fix as input files
- Click OK button (See Screen Capture for CALD Parameterization (Voxel Surface))
Surface Visualization (ADC_ParaMap, Mesh)¶
Task
Visualize surface meshes in both object and parameter spaces
Input
SpharmMatDir/data/Ex0302/hip03_smo/*_smo.mat or
SpharmMatDir/data/Ex0302/hip03_smo/initParamCALD/*_ini.mat
Output
SpharmMatDir/data/Ex0302/hip03_smo/PNG/*.png or
SpharmMatDir/data/Ex0302/hip03_smo/initParamCALD/PNG/*.png
Steps
- Select DisplayObjs under the Utils pop-up menu (bottom-left corner)
- In the right panel, let Space be both, Mesh be orig, Shade be mesh, Overlay be adc_paramap, Export be PNG
- In the right panel, click ..... button next to Select Input, and select either all the SpharmMatDir/data/Ex0302/hip03_smo/PNG/*.png files or all the SpharmMatDir/data/Ex0302/hip03_smo/initParamCALD/PNG/*.png as the input files
- Click OK button (See Screen Capture for Surface Visualization (ADC_ParaMap, Mesh))
Notes
- The visualization results are saved as PNG files under SpharmMatDir/data/Ex0302/hip03_smo/PNG and SpharmMatDir/data/Ex0302/hip03_smo/initParamCALD/PNG.
- The results of initial parameterization and optimized parameterization for a hippocampus are shown in Surface Visualization (ADC_ParaMap, Mesh), where the ADC (area distortion cost) is defined in Area Distortion Cost.
Exercise 3.3 Voxel Surfaces (PDM)¶
This exercise was tested on a WinXP machine (3GHz CPU, 3.25G RAM) running Matlab 7.7.0 (R2008b) and SPHARM-PDM (WinXP-v1.3). It took 30-40 minutes to parameterize all 40 objects.
Major Steps
- PDM Parameterization for Voxel Surfaces
- Format Conversion surf_para_meta2smo
- Surface Visualization (ADC_ParaMap, Solid with Mesh)
PDM Parameterization for Voxel Surfaces¶
Task
Create spherical parameterization for voxel surfaces using SPHARM-PDM
Input
SpharmMatDir/data/Ex0303/hip03_gipl_fix/*_fix.gipl: This folder is a copy of SpharmMatDir/data/Ex0202/hip03_gipl_fix/*_fix.gipl. The surface of an input binary object needs to be genus zero (i.e., with spherical topology).
Output
SpharmMatDir/data/Ex0303/hip04_meta/*.meta: Object and parameter meshes are stored in *_surf.meta and *_para.meta respectively.
Steps
- Make an output directory SpharmMatDir/data/Ex0303/hip04_meta
- Run SPHARM_MAT.m under Matlab
- Click Parameterization button
- In the right panel, let Method be PDM
- In the right panel, let iter be 500, label be 1, others be empty.
- In the right panel, select SpharmMatDir/data/Ex0303/hip04_meta as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *_fix.gipl files under SpharmMatDir/data/Ex0303/hip03_gipl_fix as input files
- Click OK button (See Screen Capture for PDM Parameterization (Voxel Surface))
Format Conversion surf_para_meta2smo¶
Task
Convert META surface format generated by SPHARM-PDM to matlab format
Input
SpharmMatDir/data/Ex0303/hip04_meta/*.meta
Output
SpharmMatDir/data/Ex0303/hip05_smo/*_smo.mat
Steps
- Make an output directory SpharmMatDir/data/Ex0303/hip05_smo/
- Run SPHARM_MAT.m under Matlab
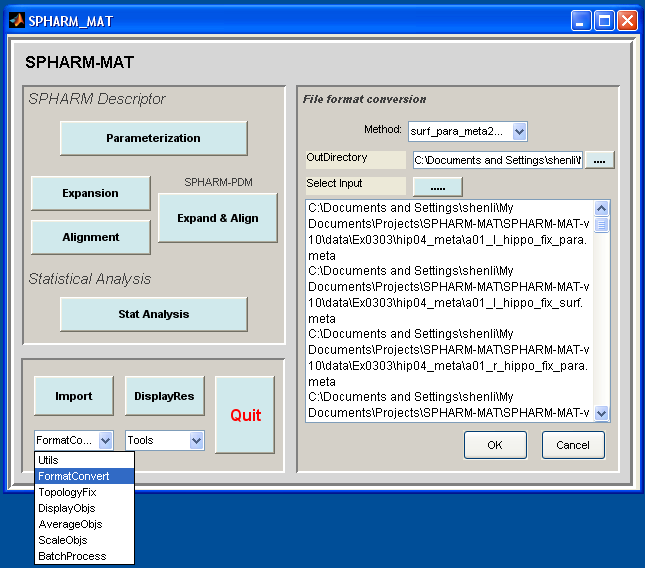
- Select FormatConvert under the Utils pop-up menu (bottom-left corner)
- In the right panel, let Method be surf_para_meta2smo
- In the right panel, select SpharmMatDir/data/Ex0303/hip05_smo/ as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *.meta files under SpharmMatDir/data/Ex0303/hip04_meta/ as input files
- Click OK button (See Screen shot for Format Conversion surf_para_meta2smo.)
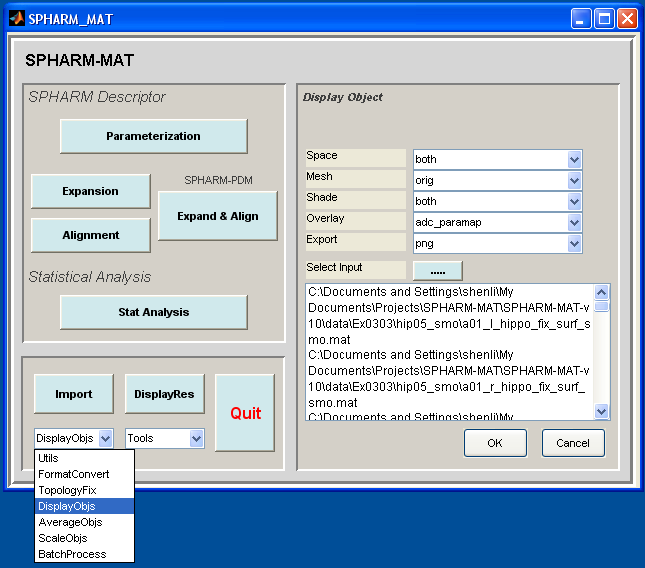
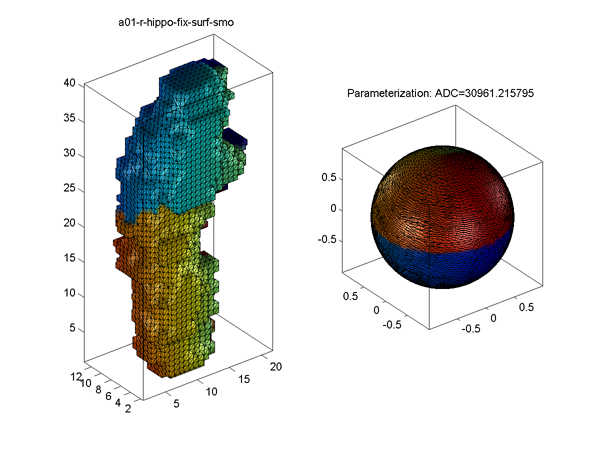
Surface Visualization (ADC_ParaMap, Solid with Mesh)¶
Task
Visualize surface meshes in both object and parameter spaces
Input
SpharmMatDir/data/Ex0303/hip05_smo/*_smo.mat
Output
SpharmMatDir/data/Ex0303/hip05_smo/PNG/*.png
Steps
- Select DisplayObjs under the Utils pop-up menu (bottom-left corner)
- In the right panel, let Space be both, Mesh be orig, Shade be both, Overlay be adc_paramap, Export be PNG
- In the right panel, click ..... button next to Select Input, and select all the SpharmMatDir/data/Ex0303/hip05_smo/PNG/*.png files as the input files
- Click OK button (See Screen Capture for Surface Visualization (ADC_ParaMap, Solid with Mesh))
Notes
- The visualization results are saved as PNG files under SpharmMatDir/data/Ex0303/hip05_smo/PNG.
- The parameterization result for a hippocampus is shown in Surface Visualization (ADC_ParaMap, Solid with Mesh), where the ADC (area distortion cost) is defined in Area Distortion Cost.
- To understand why ADC is so high in this example, see ADC Calculation for Voxel Surface and Its Triangulation.
Relevant Information¶
Useful Tips