Introduction¶
License¶
SPHARM-MAT uses GNU General Public License (GPL). The license description is included in the software package. Please review and accept the license before installing SPHARM-MAT via any source.
Download and Installation¶
Software
Available at http://www.iu.edu/~spharm/.
Documentation
Install SPHARM-MAT¶
- Download
- The SPHARM-MAT package is available at http://www.iu.edu/~spharm/.
- Install
Unzip the package and move the folder to an appropriate place, denoted as SpharmMatDir in the rest of the documentation. There are three subfolders under SpharmMatDir:
- SpharmMatDir/code/: SPHARM-MAT code
- SpharmMatDir/code/license: License description
- SpharmMatDir/data/: Test data used in the tutorial
- SpharmMatDir/scripts/: Command line scripts for running all the exercises under Matlab
- Launch
- To start SPHARM-MAT, run SpharmMatDir/code/SPHARM_MAT.m under Matlab.
- Build C mex (optional)
- The released package includes the precompiled mex files (“LocalSmoothing” and “simple”). However, they may not work on different computing environment from where they were compiled. Therefore, to make sure that they work on your computer, you can compile LocalSmoothing.c and simple.c, located in SpharmMatDir/code/C_sources and copy the mex files to SpharmMatDir/code.
How to compile simple.c
On any platform
mex simple.c
How to compile LocalSmoothing.c
If you are using the Lcc compiler that ships with MATLAB on MS Windows OS:
mex LocalSmoothing.c matlabroot/extern/lib/win32/lcc/libmwlapack.lib matlabroot/extern/lib/win32/lcc/libmwblas.lib
If you are using Microsoft Visual C++ as your C compiler on MS Windows OS:
mex LocalSmoothing.c matlabroot/extern/lib/win32/microsoft/libmwlapack.lib matlabroot/extern/lib/win32/microsoft/libmwblas.lib
or
mex LocalSmoothing.c matlabroot/extern/lib/win64/microsoft/libmwlapack.lib matlabroot/extern/lib/win64/microsoft/libmwblas.lib
On other platform:
mex LocalSmoothing.c -lmwlapack -lmwblas
Install SPHARM-PDM¶
- Recommended
- It is highly recommended to install SPHARM-PDM if you want to process voxel surfaces. Be aware of the difference in area distortion calculation between a voxel surface and its corresponding triangulation; see ADC Calculation for Voxel Surface and Its Triangulation.
- Download
- SPHARM-PDM is available at http://www.nitrc.org/projects/spharm-pdm
Configure SPHARM-MAT for connecting it to SPHARM-PDM.
- Run SPHARM_MAT.m
- Select Tools->Config
- In the “Configuration” window, set “PDM_path_pc” or “PDM_path_unix”
- You only need to do this ONCE unless the PDM_path changes
- You should make sure that SPHARM-PDM is working correctly
SPHARM-MAT Overview¶
SPHARM Method¶
SPHARM-MAT is implemented based on a powerful 3D Fourier surface representation method called SPHARM [Brechb1995], which creates parametric surface models using spherical harmonics. Spherical harmonics were first used as a type of parametric surface representation for radial or stellar surfaces 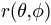 . An extended method, called SPHARM, was proposed by [Brechb1995] to model more general shapes, where three functions of
. An extended method, called SPHARM, was proposed by [Brechb1995] to model more general shapes, where three functions of 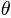 and
and 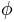 were used to represent a surface. SPHARM is a widely used shape description method for arbitrarily shaped but simply connected 3D objects. It is suitable for surface comparison and can deal with protrusions and intrusions. It is a fine-scale global descriptor with several advantages, including inherent interpolation, accurate scaling, and implicit correspondence. It can be used to derive other widely used shape descriptors such as landmarks, deformation fields, and medical axes. In addition, SPHARM has been successfully applied to many applications in medical imaging.
were used to represent a surface. SPHARM is a widely used shape description method for arbitrarily shaped but simply connected 3D objects. It is suitable for surface comparison and can deal with protrusions and intrusions. It is a fine-scale global descriptor with several advantages, including inherent interpolation, accurate scaling, and implicit correspondence. It can be used to derive other widely used shape descriptors such as landmarks, deformation fields, and medical axes. In addition, SPHARM has been successfully applied to many applications in medical imaging.
SPHARM is essentially a Fourier transform technique that defines a 3D surface using three spherical functions and transforms them into three sets of Fourier coefficients in the frequency domain. Three steps are involved to obtain a SPHARM shape descriptor:
- Spherical parameterization creates a continuous and uniform mapping from the object surface to the surface of a unit sphere, and its result is a bijective mapping between each point
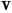 on a surface and a pair of spherical coordinates
on a surface and a pair of spherical coordinates 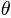 and
and 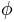 :
: 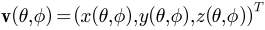 .
. - SPHARM expansion expands the object surface into a complete set of spherical harmonic basis functions
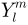 , where
, where 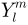 denotes the spherical harmonic of degree
denotes the spherical harmonic of degree 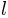 and order
and order 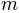 and it is essentially a Fourier basis function defined on the sphere. The expansion takes the form:
and it is essentially a Fourier basis function defined on the sphere. The expansion takes the form: 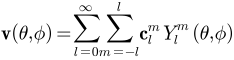 , where
, where 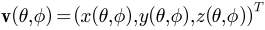 and
and 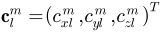 . The Fourier coefficients
. The Fourier coefficients 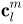 up to a user-desired degree can be estimated by solving a linear system. The object surface can be reconstructed using these coefficients, and using more coefficients leads to a more detailed reconstruction.
up to a user-desired degree can be estimated by solving a linear system. The object surface can be reconstructed using these coefficients, and using more coefficients leads to a more detailed reconstruction. - SPHARM alignment creates a shape descriptor (i.e., excluding translation, rotation, and scaling) from a normalized set of SPHARM coefficients, which are comparable across objects. The first two steps are necessary for modeling an individual shape, while the third step is mainly for group analysis.
SPHARM-MAT, SPHARM-PDM, SurfStat¶
SPHARM-PDM [Styner2006] is an existing SPHARM shape analysis tool and is a part of the NA-MIC toolkit. SPHARM-MAT (SPHARM Modeling and Analysis Toolkit) is a synergistic effort to SPHARM-PDM. While SPHARM-PDM is implemented in C/C++, SPHARM-MAT is Matlab-based. The advantage of having an additional tool on a different platform is twofold: (1) flexibility for users to choose the platform they favor, and (2) opportunity for tool comparison and cross-validation. Compared with SPHARM-PDM, SPHARM-MAT has a few additional features summarized below.
- Spherical parameterization: SPHARM-MAT implements the conventional spherical parameterization procedure proposed in [Brechb1995], which can be applied only to voxel surfaces. However, 3D surfaces are in general represented as triangular meshes. SPHARM-MAT implements a spherical parameterization algorithm that makes the SPHARM model applicable to general triangle meshes.
- SPHARM expansion: The SPHARM coefficients computed by SPHARM-MAT are complex numbers. The SPHARM coefficients computed by SPHARM-PDM are real numbers, where the imaginary part of each coefficient is ignored.
- SPHARM alignment: Typical SPHARM registration implemented in SPHARM-PDM uses the first order ellipsoid (FOE) for establishing surface correspondence and aligning objects, which may not be sufficient in some cases. In addition to FOE alignment, SPHARM-MAT implements SHREC, an efficient and general-purpose surface matching method for registering 3D SPHARM models.
SPHARM-MAT includes a few components (in both GUI and command line) that interface with SPHARM-PDM. It is highly recommended to install SPHARM-PDM if you want to parameterize voxel surfaces.
SPHARM-MAT contains two simple statistical components for group analysis. For more complicated statistical models, we suggest that SurfStat [Worsley2008] should be considered. Surfstat is a free software tool which performs statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. Since SurfStat is also matlab-based, it is straightforward to integrate SurfStat with SPHARM-MAT for statistical surface analysis. For example, one can first use SPHARM-MAT to model and align surfaces and extract surface signals, and then use SurfStat for statistical inference on the surface.
Documentation Overview¶
Tutorial and Processing Pipeline¶
A set of exercises are provided in the following six chapters to serve as a tutorial for showing how to use SPHARM-MAT. While the last chapter shows a few miscellaneous functions, the first five chapters naturally form the processing pipeline of SPHARM-MAT.
- Data Preparation: Preprocess 3D binary image data.
- Spherical Parameterization: Map object surface onto sphere with distortion minimization.
- SPHARM Expansion: Expand the surface into spherical harmonic representation.
- Surface Alignment: Align all SPHARM models together for subsequent group analysis.
- Statistical Analysis: Perform statistical shape analysis.
- Other Utilities: Demonstrate a few other SPHARM-MAT utilities.
After completing all the exercises in these chapters, you should have a basic idea on how SPHARM-MAT works.
Command Line and Batch Processing¶
All the exercises are designed for using the graphical user interface (GUI). However, for each exercise, we have put together a command line script in SpharmMatDir/scripts/. Running this script completes the same task in the batch mode. So if you don’t want to work with the GUI, you can modify these command line scripts to form your own batch processing task.
System Features¶
Besides the tutorial, we also provide the following chapters with system relevant features.
- System Components and Features: Organized based on GUI components.
- Tips: Useful tips.
- Changes: Keep track of changes.
- Bibliography: Reference information.
Data Structure¶
We summarize our naming convention for various data files as follows.
*_bim.mat: 3D binary objects
- bim: binary image
- origin: coordinates of voxel (1,1,1)
- vxsize: voxel size
*_fix.mat: 3D binary objects after topology fix
- bim: binary image
- origin: coordinates of voxel (1,1,1)
- vxsize: voxel size
*_obj.mat: original surface meshes
- vertices: vertices on the surface
- faces: mesh elements
*_ini.mat: surface mesh after initial parameterization
- vertices: vertices on the surface
- faces: mesh elements
- sph_verts: spherical mapping of vertices
- dateline: date line on the sphere (
 )
) - landmarks: vertices of north, south, east, and west on the sphere
*_smo.mat: surface mesh after optimized parameterization
- vertices: vertices on the surface
- faces: mesh elements
- sph_verts: spherical mapping of vertices
- measure: distortion measures of the parameterization
*_des.mat: surface mesh with SPHARM coefficients
- vertices: vertices on the original surface (or vertices on the SPHARM-PDM reconstruction if the file is converted from SPHARM-PDM result)
- faces: mesh elements
- sph_verts: spherical mapping of vertices
- fvec: SPHARM coefficients (complex numbers)
*_prm.mat: surface mesh with SPHARM coefficients after FOE alignment in the parameter space
- vertices: vertices on the surface
- faces: mesh elements
- sph_verts: spherical mapping of vertices
- fvec: SPHARM coefficients (complex numbers)
*_reg.mat: surface mesh with SPHARM coefficients after registration
- vertices: vertices on the registered surface (not available for SHREC results)
- faces: mesh elements (not available for SHREC results)
- sph_verts: spherical mapping of vertices (not available for SHREC results)
- fvec: registered SPHARM coefficients (complex numbers)
t_map*.mat: Data and results for t-test
- atlas_vertices: vertices on the atlas surface
- faces: mesh elements
- sph_verts: spherical mapping of atlas_vertices
- grInfo: group ID for each subject
- groupID: annotation for each group ID
- pvalue: vertex-wise p values
- tstats: vertex-wise t values
- signal: description of the signal
- vtnorm: surface normals
- FWHM: size of heat kernel used for surface signal smoothing
PCA_stat*.mat: Data and results for PCA analysis
- eigenvals: eigenvalues
- eigenvecs: eigenvectors
- fvecs: a set of SPHARM coefficients
- groupID: group ID
- scores: principal component (PC) scores
- perc_variance_explained: percentage of data variance explained by each PC
- cum_percent_explained: cumulated percentage of data variance explained by the first n PCs
*.gipl: 3D binary objects in GIPL format
- Used by SPHARM-PDM: see Exercise 2.2: SPHARM-PDM Topology Fix
- See also Format Conversion for Binary Objects
*.meta: surface mesh in META format
- Used by SPHARM-PDM: see Exercise 3.3 Voxel Surfaces (PDM)
- See also Format Conversion for Surface Meshes
- See also Format Conversion for SPHARM Models
*.coef: SPHARM coefficients (real parts kept, imaginary parts ignored)
- Used by SPHARM-PDM: see Exercise 4.2 SPHARM-PDM Expansion
- See also Format Conversion for SPHARM Models
Team and Contact¶
The first release of SPHARM-MAT was primarily supported by the following award:
NIH/NIBIB R03 EB008674 SPHARM Shape Modeling and Analysis Toolkit for Brain Imaging
PI: Li Shen, Co-PI: Andrew J. Saykin
The project was developed at:
The IU Center for Neuroimaging (CfN)
The Center for Computational Biology and Bioinformatics (CCBB)
Indiana University School of Medicine (IUSM).
The following people contributed to the development, testing and documentation of various components of SPHARM-MAT:
Major developers: Li Shen, Sungeun Kim
Other contributors: Jing Wan, John D. West, Kiernan McCullough,
Thomas Councell, Andrew J. Saykin
Collaborators at UT Arlington: Heng Huang, Fillia Makedon
Collaborator at UW Madison: Moo K. Chung
Contact:
Li Shen
Center for Neuroimaging, Dept. of Radiology and Imaging Sciences
Center for Computational Biology and Bioinformatics
Indiana University School of Medicine
950 W Walnut St, R2 E124
Indianapolis, IN 46202
Tel: (317) 278-0498
Fax: (317) 274-1067
Email: shenli@iupui.edu
URL: http://www.iupui.edu/~shenlab/
Question about SPHARM-MAT or bug reporting:
SpharmMat@gmail.com