Tips¶
This chapter provides a list of tips that address various SPHARM-MAT issues, including strengths, limitations, parameter settings, options, etc.
File Selection¶
Matlab starts navigation for file selection from the current directory. Thus, to minimize the effort of finding your data, you can change matlab current directory to your data directory if you want to work on the same data for a while.
ADC Calculation for Voxel Surface and Its Triangulation¶
We notice that the ADC values of SPHARM-PDM results are often extremely high; see Surface Visualization (ADC_ParaMap, Solid with Mesh) for an example. Some relevant discussion is provided below.
- First of all, the spherical parameterization created by SPHARM-PDM is a very high quality area preserving mapping for voxel surfaces, where each square face is mapped to a spherical quadrilateral with the same relative area.
- The major reason here is that our area distortion cost is calculated based on triangles instead of quadrilaterals. SPHARM-PDM creates an equal area mapping between quadrilaterals on the object space and parameter space. However, this area preserving property no longer holds for a triangulation of the same surface, where the triangulation is created by splitting each quadrilateral into two triangles. On the object surface, each quadrilateral (a square for the voxel surface case) always splits into two triangles with the same area, while this is not true on the parameter surface. Consequently, higher ADC values are generated from triangulation-based calculation for area distortion.
- In particular, for very slim quadrilaterals on the sphere, if the long diagonal happens to be selected for generating triangles, some of these triangles might have an area close to zero or even flipped. In these cases, very high ADCs are introduced.
- As mentioned in [Quicken2000], imposing constraints on area preservation results in only two degrees of freedom for distortion minimization for triangular meshes. However, for quadrilateral meshes, there are n degrees of freedom available, where n is the number of vertices on the surface. As a result, in terms of spherical parameterization, it is much more difficult to generate an area preserving map for a triangular mesh than a quadrilateral mesh of the same surface. This is another reason why an equal area mapping for a quadrilateral mesh does not imply an equal area mapping for the corresponding triangulation.
- Finally, even though the ADCs are high, SPHARM-PDM is still highly recommended for parameterizing voxel surfaces. Based on visual inspection, the PDM results usually preserve the area better than CALD ones; see Surface Visualization (icosa4, Solid with Mesh) and Surface Visualization (quad64, Mesh).
Parameterization CALD Input and Output¶
Input
Parameterization using CALD as Method can take a binary object, a triangular mesh, or a quadrilateral mesh as an input. For a binary object, SPHARM-MAT extracts its voxel surface and describes it as a triangular mesh. For a quadrilateral mesh, SPHARM-MAT converts it to a triangular mesh. In any case, the resulting triangular mesh needs to be a genus zero surface (i.e., with a spherical topology). If you work with binary images, be sure to run topology fix first (see Data Preparation).
Output
Results of both initial parameterization and optimized parameterization are saved. See Surface Visualization (ADC_ParaMap, Solid with Mesh) for an example.
Parameterization CALD Parameters¶
Spherical parameterization using CALD has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 3.1 Surface Meshes (CALD) and Exercise 3.2 Voxel Surfaces (CALD).
- MeshGridSize: Used in global smoothing for creating regular meshes
- MaxSPHARMDegree: Used in global smoothing for interpolating the area scaling ratio functions
- Tolerance: Used in global smoothing for reducing extreme values
- Smoothing: Used in global smoothing for smoothing the area scaling ratio functions
- Iteration: Max number of iterations for global smoothing
- LocalIteration: Number of iterations in each local smoothing step
- t_major: x major smooths latitude first and longitude second, y major does the opposite. This option was used in a previous implementation. It should be ignored in the current release.
- SelectDiagonal: Used for converting quadrilaterals to triangles. ShortDiag uses the short diagonal to split the quadrilateral, LongDiag uses the long one for splitting. This option was used in a previous implementation. It should be ignored in the current release.
Parameterization PDM Input¶
Parameterization using PDM as Method takes a binary object as input. The surface of the binary object needs to be genus zero (i.e., with a spherical topology). To meet this requirement, be sure to run topology fix first (see Data Preparation).
Parameterization PDM Parameters¶
Parameterization using PDM as Method has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 3.3 Voxel Surfaces (PDM).
See also
SPHARM-PDM manual for further details about these parameters.
iter: Max number of iterations (default 500)
label: Binary volume is formed by all the voxels with value equal to the specified label (default 1)
- others: Other command line parameters used in SPHARM-PDM
- -outbase <basefile>: base filename for output
- -initParam meshfile: initialization of parameterization, file has to be a mesh of same triangle topology as the one extracted from the image
- -v: verbose mode
- -vxml: Display an XML description on the standard output
SPHARM Expansion Degree¶
The SPHARM coefficients 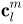 up to a user-desired degree are computed in SPHARM expansion step. The object surface can be reconstructed using these coefficients, and using more coefficients leads to a more detailed reconstruction; see Surface Visualization (icosa4, Solid with Mesh).
up to a user-desired degree are computed in SPHARM expansion step. The object surface can be reconstructed using these coefficients, and using more coefficients leads to a more detailed reconstruction; see Surface Visualization (icosa4, Solid with Mesh).
SPHARM Expansion has the following parameter for users to specify the maximum degree; see Exercise 4.1 SPHARM-MAT Expansion.
- MaxSPHARMDegree: SPHARM coefficients up to this degree are stored in the model
SPHARM Expansion PDM Difference¶
The SPHARM coefficients computed by SPHARM-MAT (see Exercise 4.1 SPHARM-MAT Expansion) are complex numbers. The SPHARM coefficients computed by SPHARM-PDM (see Exercise 4.2 SPHARM-PDM Expansion) are real numbers, where the imaginary part of each coefficient is ignored.
Alignment FOE Parameters¶
Alignment using FOE has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 5.1 FOE Alignment.
- CPoint {x/y/z}: CPoint is the crossing point of equator and zero meridian it should be on the positive side of x, y, or z axis in the object space
- NPole {x/y/z}: NPole is the north pole and it should be on the positive side of x, y, or z axis in the object space
- MaxSPHARMDegree: SPHARM coefficients up to this degree are stored in the results
Alignment SHREC Parameters¶
Alignment using SHREC has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 5.2 SHREC Alignment.
- Template: To which all the SPHARM models are registered
- MaxSPHARMDegree: SPHARM coefficients up to this degree are stored in the results
- GroupAlpha: Number of rotations to be processed together (i.e., matrix operation instead of loop)
- NormalizeSize (Yes/No): If yes, do the scaling for normalizing the centroid size
- BaseRes: Base resolution of icosahedral subdivision to hierarchically sample the Euler rotation angles from icosahedral mesh.
- HierarchyStep: Step of hierarchical sampling scheme of Euler rotation angles (use 1 for now, >1 seems to be very slow, improvement needed).
- HiderarchyDepth: Depth of hierarchical sampling scheme of Euler rotation angles.
- Top_K: Number of selected angles at each iteration.
- GammaRes: Sampling resolution of gamma angles.
Alignment SHREC Initial Models¶
Initial SPHARM models for SHREC alignment: If the FOE of your data is a real ellipsoid having three distinct axes (e.g., like hippocampus), you may want to run FOE alignment first before doing SHREC. Note that the final results of FOE alignment do not preserve the original geometric information in the object space. Therefore, be sure to apply SHREC only to the intermediate results (i.e., *_prm.mat files in Exercise 5.2 SHREC Alignment) of FOE alignment. For these intermediate results, the orientation and the location of the objects have not been transformed in the object space; however, their parameter nets have been aligned to the canonical position so that an initial surface correspondence has been established among then.
Alignment SHREC Template¶
In SHREC alignment, a template needs to be specified so that all the individual SPHARM models can be registered to it. This template should be carefully picked or prepared. For a case-control group study, it could be an individual control object or the mean shape of a set of typical control objects (see Exercise 7.2 Average Objects). In addition, its underlying parameterization should be oriented to a canonical position. If the FOE of the template is a real ellipsoid, FOE alignment can be applied to achieve this goal, as described in Alignment SHREC Initial Models; otherwise, manual adjustment could be used to rotate the parameter net to a user-desired position.
Expand and Align Parameters¶
The Expand & Align button is provided for performing SPHARM expansion and FOE alignment using SPHARM-PDM. It has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 4.2 SPHARM-PDM Expansion and Exercise 5.3 SPHARM-PDM FOE Alignment.
See also
SPHARM-PDM manual for further details about these parameters.
flipTemplate: Set a fliptemplate for normalization of axis-flips (could be empty)
subdivLevel: Set subdivision level for linear icosahedron subdivision
spharmDegree: Set the degree of spherical harmonic expansion
regTemplate: Specify a registration template (could be empty)
- FinalFlip: Allows an optional additional flipping of the parameterization
- This is rarely necessary, only in case the general first order ellipsoid heuristic fails. Flipping of the parameterization ALONG a given axis will lead to a flip of the coordinates ALONG the same axis in the ellipse aligned mesh
- There are 7 possibilities (1..7, 0 = no flip) of parameterization flipping along an axis
- 1 = flip along axes of x & y, 2 = flip along y & z, 3 = flip along x & z, 4 = flip along x, 5 = flip along y, 6 = flip along x & y & z, 7 = flip along z, where y is the smallest, x is the second smallest and z is the long axis of the ellipsoid
- Others:
- -outbase <basefile>: Set base filename for output (*_surfSPHARM.coef, *_surfSPHARM.meta, *_surfSPHARM_ellalign.coef, *_surfSPHARM_ellalign.meta, *_surfSPHARM_procalign.meta)
- -flipRegTemplate <meshfile>: Set a template for parameter normalization and registration
- -flipRegPoints <points.samp>: Set sample points on flipRegTemplate for parameter alignment
- -paraOut: Write sphere parameterization and coloring to file
- -NoParaAlign: Do not do any alignment/correspondence of the parameterization using the first order ellipsoid
- -v: Verbose mode
- -vxml: Display an XML description on the standard output
Stat Analysis T Map Parameters¶
Statistical analysis using t-map as method has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 6.1 T Test.
- Atlas: The template surface used for extracting surface signals and for visualizing results
- Smoothing_FWHM: Size of kernel for surface signal smoothing using heat kernel method (see [Chung2005])
- EqualVariance: Equal variance assumption for t-test
- Signal: vl_defm_org, length of the deformation vector; vl_defm_nrm, deformation along normal direction of the atlas; vl_defm_pca, deformation component along PCA direction; vl_defm_fld, deformation component along FLD direction; see [Shen2006b] for details
- SampleMesh: Underlying mesh for SPHARM reconstruction
- OutputNamePrefix: Prefix of the name of the output statistics file
- GroupIDs: Group 1 ID and Group 2 ID separated by comma
Stat Analysis PCA Parameters¶
Statistical analysis using PCA as method has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 6.2 PCA.
- GroupID: Group ID for selected objects
- OutputName: File name of output statistics
DisplayRes T-Map Parameters¶
DisplayRes using res-t-map as method has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 6.1 T Test.
- Threshold_p_value: Regions with uncorrected p<threshold_p_value are color-mapped
- Overlay: Selection of p-value or t-map for display.
- Colormap: Selection of colormap, jet/hot/summer/cool/autumn/winter/spring.
DisplayRes PCA Parameters¶
DisplayRes using res-PCA as method has a few adjustable parameters shown below. In most cases, you can just use the default setting; see Exercise 6.2 PCA.
- Level: Number of principal components to display
- Sigma: Number of standard deviations in each principal component axis to display
- Mesh: Underlying mesh for SPHARM reconstruction, quad32/quad64/../quad512/icosa1/.../icosa6
- MaxSPHARMDegree: Maximum degree of SPHARM coefficients used for reconstruction
Import Parameters¶
Import utility does the following conversion (see Exercise 7.1 Import)
- Read in binary images in nifti and/or analyze formats and convert them to *_bim.mat files
- Read in surface meshes in stl and/or Amira matlab formats and convert them to *_obj.mat files
Import has the following parameter to adjust
- ResampleFactor: For upsampling (>1) or downsampling (<1) the input binary volumes (nifti and analyze); no effect on surface meshes (stl and Amira matlab).
Format Conversion for Binary Objects¶
SPHARM-MAT uses matlab format to store 3D binary objects (before topology fix: *_bim.mat; after topology fix: *_fix.mat). SPHARM-PDM uses gipl format to store 3D binary objects (*.gipl). FormatConvert under the Utils pop-up menu can be used for performing format conversion with the following three options:
- bim2gipl: Convert *_bim.mat to *.gipl; see Screen shot for Format Conversion bim2gipl..
- fix2gipl: Convert *_fix.mat to *.gipl.
- gipl2bim: Convert *.gipl to *_bim.mat; see Screen shot for Format Conversion gipl2bim.
Format Conversion for Surface Meshes¶
SPHARM-MAT uses matlab format to store surfaces (original: *_obj.mat; after initial parameterization: *_fix.mat; after parameterization optimization: *_smo.mat). SPHARM-PDM uses META format to store object meshes (*_surf.meta) and parameter meshes (*_para.meta). FormatConvert under the Utils pop-up menu can be used for performing conversion between these formats:
- surf_para_meta2smo: Convert *.meta to *_smo.mat; see Screen shot for Format Conversion surf_para_meta2smo..
- smo2surf_para_meta: Convert *_smo.mat to *.meta.
Format Conversion for SPHARM Models¶
SPHARM-MAT uses matlab format to store SPHARM models:
- in the original coordinate system: *_des.mat
- in the FOE/SHREC aligned coordinate system: *_reg.mat
SPHARM-PDM uses META format (i.e., surface meshes) and COEF format (i.e., SPHARM coefficients) to store SPHARM models:
- in the original coordinate system: *_surfSPHARM.meta and *_surfSPHARM.coef
- in the FOE aligned coordinate system: *_surfSPHARM_ellalign.meta and *_surfSPHARM_ellalign.coef
- in the Procrustes aligned coordinate system: *_surfSPHARM_procalign.meta (only reconstructed surface meshes available)
FormatConvert under the Utils pop-up menu can be used for performing conversion between these formats:
- meta_coef2des: Convert *_surfSPHARM.meta and *_surfSPHARM.coef to *_des.mat; see Screen shot for Format Conversion meta_coef2des..
- des2meta_coef: Convert *_des.mat to *.meta and *.coef.
- ellalign_meta_coef2reg: Convert *_surfSPHARM_ellalign.meta and *_surfSPHARM_ellalign.coef to *_reg.mat; see Screen shot for Format Conversion ellalign_meta_coef2reg..
- reg2proalign_meta: Convert *_reg.mat to *_proalign.meta and *.coef.
TopologyFix InHouse_Fix Option¶
TopologyFix under the Utils pop-up menu has two options for customizing 3D hole filing process if Inhouse_Fix is selected as Method:
- Connectivity: Neighborhood connectivity can be defined by one of the four options 1=(6+,18), 2=(18,6+), 3=(6,26), 4=(26,6). More details are available in http://dx.doi.org/10.1016/S0167-8655(01)00152-0.
- Epsilon: Hole size can be specified by voxel count so that holes bigger than Epsilon won’t be filled.
TopologyFix PDM_Fix Option¶
TopologyFix under the Utils pop-up menu has two options for customizing 3D hole filing process if PDM_Fix is selected as Method:
- space: You can define voxel size and SPHARM-PDM will re-slice the volume.
- other: You can define other command line options used by SPHARM-PDM.
DisplayObjs Space Option¶
DisplayObjs under the Utils pop-up menu has three options for selection of visualizing the surface in object and/or parameter spaces:
- object: A surface is rendered in the object space (e.g., Binary Object Visualization (Solid with Mesh)).
- param: A surface is rendered in the parameter space.
- both: A surface is rendered in both object and parameter spaces (e.g., Surface Visualization (ADC_ParaMap, Solid with Mesh)).
DisplayObjs Mesh Option¶
DisplayObjs under the Utils pop-up menu has a few options for selecting mesh structures:
- orig: A original surface mesh is selected for rendering (e.g., Binary Object Visualization (Solid with Mesh)).
- quad32-quad512: A SPHARM reconstruction based on a regular 32-by-32, ... , or 512-by-512 mesh is rendered.
- icosa1-icosa6: A SPHARM reconstruction based on an icosahedral mesh at level 1, ... , or 6 is rendered.
DisplayObjs Shade Option¶
DisplayObjs under the Utils pop-up menu has three options for shading the surface:
- solid: A solid surface is rendered (e.g., Screen Capture for Surface Visualization (ADC_ParaMap, Solid with Mesh)).
- mesh: A surface mesh is rendered (e.g., Binary Object Visualization (Mesh)).
- both: A solid surface with mesh is rendered (e.g., Binary Object Visualization (Solid with Mesh)).
DisplayObjs Overlay Option¶
DisplayObjs under the Utils pop-up menu has an option for showing additional information
none: No additional information is shown.
- adc_paramap: This option has two functions.
- If Space is set as both, the ADC (see Area Distortion Cost) will be calculated and displayed.
- If Shade is set as solid or both, a color map will be displayed on the surface to visualize the underlying parameterization. For north hemisphere (i.e.,
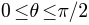 ), the color goes from green (i.e.,
), the color goes from green (i.e., 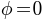 ) to red (i.e.,
) to red (i.e., 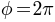 ); for south hemisphere (i.e.,
); for south hemisphere (i.e., 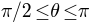 ), the color goes from green (i.e.,
), the color goes from green (i.e., 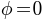 ) to blue (i.e.,
) to blue (i.e., 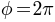 ).
).
- The option could be used in the following scenarios.
- To check the quality of spherical parameterization: See Surface Visualization (ADC_ParaMap, Solid with Mesh), Surface Visualization (ADC_ParaMap, Mesh), Screen Capture for Surface Visualization (ADC_ParaMap, Solid with Mesh) for a few examples.
- To visualize the correspondence between SPHARM models: See Surface Visualization (quad64, Degree 1), Surface Visualization (quad64, template), Screen Capture for Surface Visualization (quad64) for a few examples.
DisplayObjs Export Option¶
DisplayObjs under the Utils pop-up menu has three options for exporting the figure:
- screen: The figure is displayed on screen.
- png: The figure is saved as a png file.
- both: The figure is displayed on screen and saved as a png file.
DisplayObjs Degree Option¶
DisplayObjs under the Utils pop-up menu has an option called Degree for specifying the user-desired degree for SPHARM reconstruction. If empty, all the available coefficients are used for SPHARM reconstruction; otherwise, the reconstructed surface is generated only using those coefficients up to the user-specified degree. See Surface Visualization (icosa4, Solid with Mesh).
DisplayObjs Template Option¶
DisplayObjs under the Utils pop-up menu has an option called Template for specifying a template. This is often used when you want to display registered SPHARM models. If the template file exists, the root mean square distance between each input SPHARM model and the specified template model will be calculated and displayed on the z-axis. See Surface Visualization (quad64, template).
AverageObjs Parameters¶
AverageObjs under the Utils pop-up menu aims to create an average surface, which can be used as the atlas in group analysis. In a typical study, you can select a set of control objects and run this utility to create an atlas that represents an average normal surface. The method is the following: (1) let the atlas be the first model; (2) align each model to the atlas using SHREC; (3) let the atlas be the mean of all the data; and (4) repeat (2) and (3) until the atlas converges. See Surface Visualization (icosa4): for an example atlas. Once the atlas is created, you can use SHREC to register all the individual objects to the atlas (instead of using an individual object as the template, as shown in Exercise 5.2 SHREC Alignment). AverageObjs has the following parameter to adjust
- OutputName: Name of the output average surface.
ScaleObjs Parameters¶
ScaleObjs under the Utils pop-up menu can be used to scale a set of SPHARM objects. The scaling factors need to be pre-computed by users and provided as a csv file as an input file. See Exercise 7.2 Average Objects: for an example. ScaleObjs has the following parameter to adjust
- ScalingFactor: A csv file specifying a scaling factor for each input file.