Surface Alignment¶
Surface alignment is an important operation allowing for pairwise processing or group analysis across different surface models and is a critical step in many applications. The traditional SPHARM registration method ([Styner2006], [Brechb1995]) uses the first order ellipsoid (FOE) for alignment, and works when this ellipsoid is a real ellipsoid. When FOE is not a real ellipsoid, SHREC [Shen2008], a newer SPHARM registration method can be used. SHREC minimizes the mean square distance between corresponding surface parts and is designed for general cases.
In SPHARM-MAT, both FOE alignment and SHREC are available for registering SPHARM models (see Exercise 5.1 FOE Alignment and Exercise 5.2 SHREC Alignment). In addition, SPHARM-PDM can also be integrated for performing FOE alignment (see Exercise 5.3 SPHARM-PDM FOE Alignment).
Exercise 5.1 FOE Alignment¶
This exercise was tested on a WinXP machine (3GHz CPU, 3.25G RAM) running Matlab 7.7.0 (R2008b). It took a few minutes to finish.
Major Steps
- FOE Alignment
- Surface Visualization (quad64, Solid with Mesh)
FOE Alignment¶
Task
Use first order ellipsoids (FOE) to register SPHARM models so that each FOE has a canonical orientation in both object and parameter space.
Input
SpharmMatDir/data/Ex0501/hip04_des/*_des.mat: This folder is a copy of SpharmMatDir/data/Ex0401/hip04_des/*_des.mat.
Output
SpharmMatDir/data/Ex0501/hip05_reg/alignParam/*_prm.mat: intermediate results after alignment in parameter space
SpharmMatDir/data/Ex0501/hip05_reg/*_reg.mat: final results after alignment in both object and parameter space
Steps
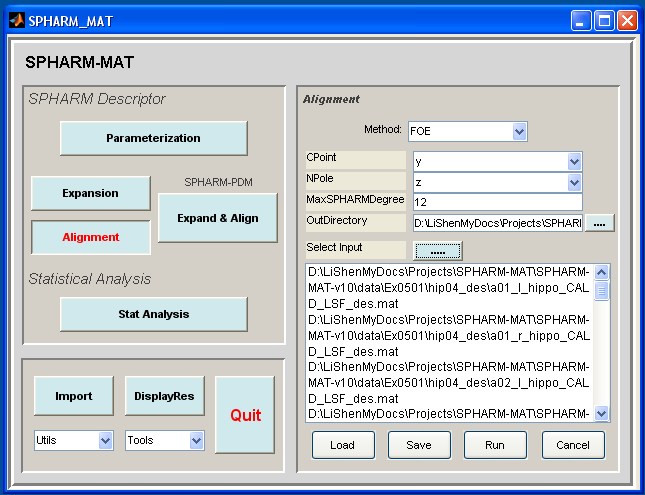
- Make an output directory SpharmMatDir/data/Ex0501/hip05_reg/
- Run SPHARM_MAT.m under Matlab
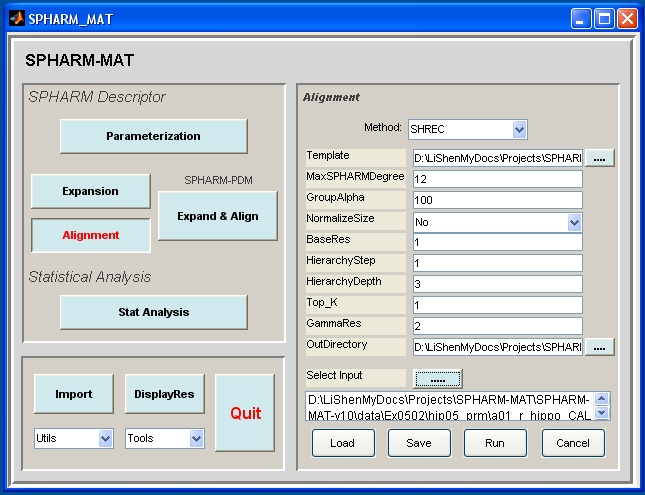
- Click Alignment button
- In the right panel, let Method be FOE
- In the right panel, let CPoint be y, NPole be z, MaxSPHARMDegree be 12
- In the right panel, select SpharmMatDir/data/Ex0501/hip05_reg/ as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *_des.mat files under SpharmMatDir/data/Ex0501/hip04_des as input files
- Click OK button (See Screen Capture for FOE Alignment)
Surface Visualization (quad64)¶
Task
Visualize SPHARM reconstruction before and after FOE alignment
Input
SpharmMatDir/data/Ex0501/hip04_des/*_des.mat
SpharmMatDir/data/Ex0501/hip05_reg/alignParam/*_prm.mat
SpharmMatDir/data/Ex0501/hip05_reg/*_reg.mat
Output
SpharmMatDir/data/Ex0501/hip04_des/PNG/*.png
SpharmMatDir/data/Ex0501/hip05_reg/alignParam/PNG/*.png
SpharmMatDir/data/Ex0501/hip05_reg/PNG/*.png
Steps
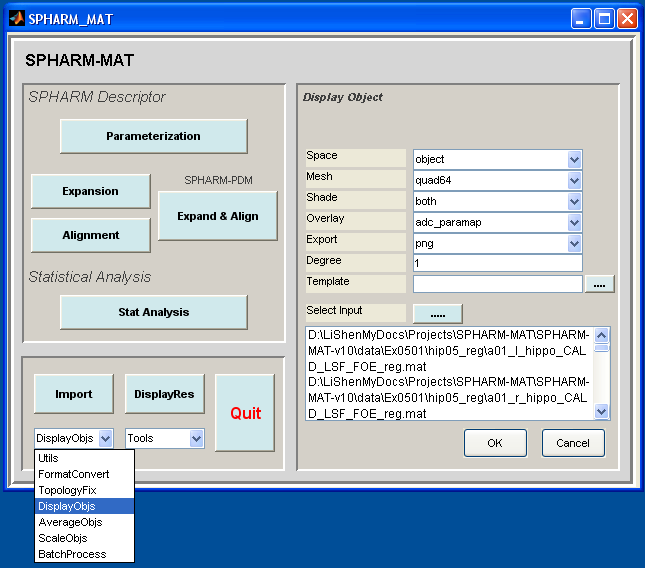
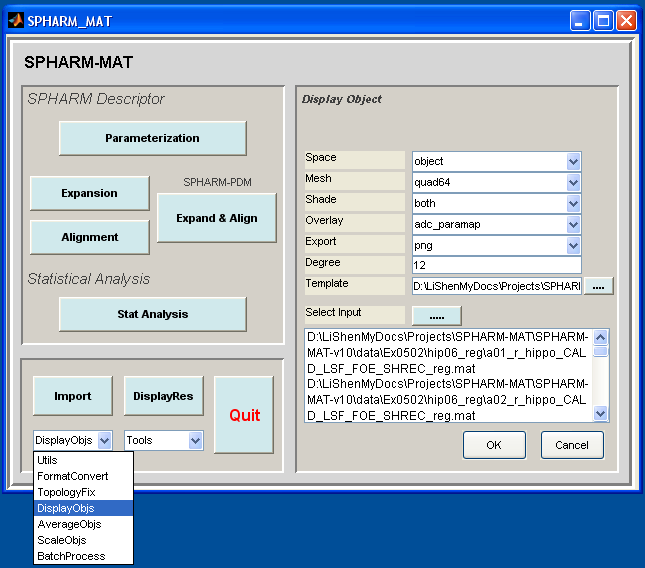
Select DisplayObjs under the Utils pop-up menu (bottom-left corner)
Be sure to run this experiments six times by setting different configurations in the right panel as follows
- Runs 1-3 (SPHARM reconstruction up to degree 1): let Space be object, Mesh be quad64, Shade be both, Overlay be adc_paramap, Export be png, Degree be 1
- Runs 4-6 (SPHARM reconstruction up to degree 12): let Space be object, Mesh be quad64, Shade be both, Overlay be adc_paramap, Export be png, Degree be 12
In the right panel, click ..... button next to Select Input, and select the following files in six different runs respectively
- Run 1 and Run 4: SpharmMatDir/data/Ex0501/hip04_des/*_des.mat
- Run 2 and Run 5: SpharmMatDir/data/Ex0501/hip05_reg/alignParam/*_prm.mat
- Run 3 and Run 6: SpharmMatDir/data/Ex0501/hip05_reg/*_reg.mat
Click OK button (See Screen Capture for Surface Visualization (quad64))
Notes
The visualization results are saved as PNG files under the PNG folder of the input directories
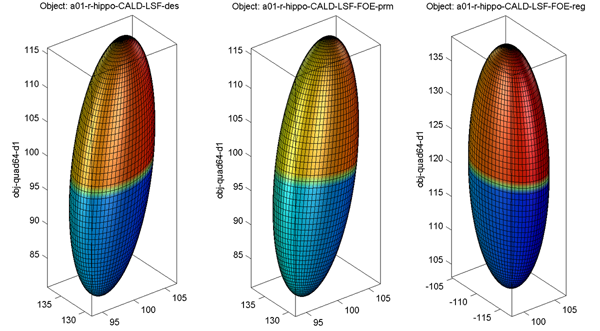
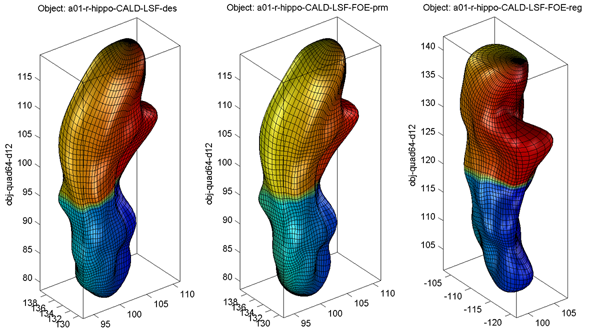
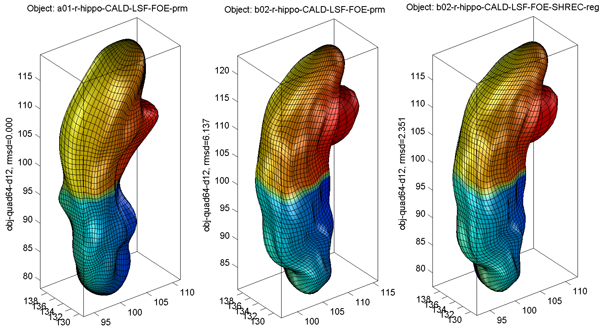
Sample alignment procedure is shown below (from left to right: original, intermediate result after aligning parameter net, final result)
- Surface Visualization (quad64, Degree 1): SPHARM reconstructions using coefficients up to degree 1 (i.e., FOE)
- Surface Visualization (quad64, Degree 12): SPHARM reconstructions using coefficients up to degree 12
See DisplayObjs Overlay Option for information about color-coded visualization of the underlying parameterization.
Relevant Information¶
Useful Tips
Exercise 5.2 SHREC Alignment¶
This exercise was tested on a WinXP machine (3GHz CPU, 3.25G RAM) running Matlab 7.7.0 (R2008b). It took about 20 minutes to finish.
Major Steps
- SHREC Alignment
- Surface Visualization (quad64, template)
SHREC Alignment¶
Task
Use SHREC to register SPHARM models to a template model for minimizing root mean square distance (RMSD)
Input
SpharmMatDir/data/Ex0502/hip05_prm/*_prm.mat: This folder is a subset of SpharmMatDir/data/Ex0501/hip05_reg/alignParam/*_prm.mat, where only right hippocampi are included.
Output
SpharmMatDir/data/Ex0502/hip06_reg/*_reg.mat
Steps
- Make an output directory SpharmMatDir/data/Ex0502/hip06_reg/
- Run SPHARM_MAT.m under Matlab
- Click Alignment button
- In the right panel, let Method be SHREC
- In the right panel, let Template be SpharmMatDir/data/Ex0502/hip05_prm/a01_r_hippo_CALD_LSF_FOE_prm.mat, MaxSPHARMDegree be 12, GroupAlpha be 100, NormalizeSize be No, BaseRes be 1, HierarchyStep be 1, HierarchyDepth be 3, Top_K be 1, GammaRes be 2
- In the right panel, select SpharmMatDir/data/Ex0502/hip06_reg/ as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *_prm.mat files under SpharmMatDir/data/Ex0502/hip05_prm/ as input files
- Click OK button (See Screen Capture for SHREC Alignment)
Notes
- See Alignment SHREC Parameters for more information about parameters.
- See Alignment SHREC Initial Models for notes about selection of initial SPHARM models.
- See Alignment SHREC Template for notes about selection of the template.
- SHREC saves only fvec in the results.
Surface Visualization (quad64, template)¶
Task
Visualize SPHARM reconstruction in object space
Input
SpharmMatDir/data/Ex0502/hip05_prm/*_prm.mat
SpharmMatDir/data/Ex0502/hip06_reg/*_reg.mat
Output
SpharmMatDir/data/Ex0502/hip05_prm/PNG/*.png
SpharmMatDir/data/Ex0502/hip06_reg/*_reg.mat/PNG/*.png
Steps
Select DisplayObjs under the Utils pop-up menu (bottom-left corner)
In the right panel, let Space be object, Mesh be quad64, Shade be both, Overlay be adc_paramap, Export be png, Degree be 12, Template be SpharmMatDir/data/Ex0502/hip05_prm/a01_r_hippo_CALD_LSF_FOE_prm.mat
In the right panel, click ..... button next to Select Input, and select the following files in two different runs respectively
- Run 1: SpharmMatDir/data/Ex0502/hip05_prm/*_prm.mat
- Run 2: SpharmMatDir/data/Ex0502/hip06_reg/*_reg.mat
Click OK button (See Screen Capture for Surface Visualization (quad64, template))
Notes
- The visualization results are saved as PNG files under the PNG folder of the input directories
- Sample alignment procedure is shown in Surface Visualization (quad64, template), where the template, the individual before alignment, and the individual after alignment are shown from left to right. The root mean square distance (RMSD) between an individual and the template is plot on the z axis.
- See DisplayObjs Template Option for information about template and RMSD.
- See DisplayObjs Overlay Option for information about color-coded visualization of the underlying parameterization.
Exercise 5.3 SPHARM-PDM FOE Alignment¶
Step 1 of this exercise was tested on a Linux machine (3GHz CPU, 8G RAM) running Matlab 7.5.0 (R2007b) and SPHARM-PDM (Linux64-v1.7), since the windows version of this function is not available. Steps 2-3 of this exercise were tested on a WinXP machine (3GHz CPU, 3.25G RAM) running Matlab 7.7.0 (R2008b) and SPHARM-PDM (WinXP-v1.3). It took a few minutes to finish.
Major Steps
- SPHARM-PDM FOE Alignment
- Format Conversion ellalign_meta_coef2reg
- Surface Visualization (quad64)
SPHARM-PDM FOE Alignment¶
Task
Use first order ellipsoids (FOE) to register SPHARM models so that each FOE has a canonical orientation in both object and parameter space.
Input
SpharmMatDir/data/Ex0503/hip04_meta/*.meta: This folder is a subset of SpharmMatDir/data/Ex0402/hip04_meta/*.meta, where only left hippocampi are included
SpharmMatDir/data/Ex0503/hip05_temp/a01_l_hippo_fix_surfSPHARM_ellalign.*: These two files are copied from SpharmMatDir/data/Ex0402/hip05_coef/ and are used as flipTemplate and regTemplate (see Expand and Align Parameters)
Output
SpharmMatDir/data/Ex0503/hip06_coef/*.*: SPHARM coefficients are stored in *.coef and SPHARM reconstructions are stored in *.meta (see also Format Conversion for SPHARM Models).
Steps
- Be sure to run this on a Linux machine, since the window version does not support this function
- Make an output directory SpharmMatDir/data/Ex0503/hip06_coef/
- Run SPHARM_MAT.m under Matlab
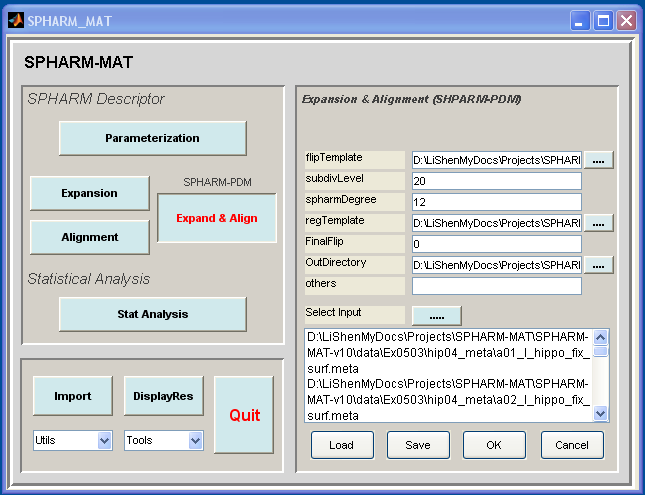
- Click Expand & Align button
- In the right panel, let flipTemplate be SpharmMatDir/data/Ex0503/hip05_temp/a01_l_hippo_fix_surfSPHARM_ellalign.coef, subdivLevel be 20, spharmDegree be 12, regTemplate be SpharmMatDir/data/Ex0503/hip05_temp/a01_l_hippo_fix_surfSPHARM_ellalign.meta, FinalFlip be 0, and other options be empty.
- In the right panel, select SpharmMatDir/data/Ex0503/hip06_coef as OutDirectory
- In the right panel, click ..... button next to Select Input, and select all the *.meta files under SpharmMatDir/data/Ex0503/hip04_meta/*.meta as input files
- Click OK button (See Screen Capture for SPHARM-PDM FOE Alignment)
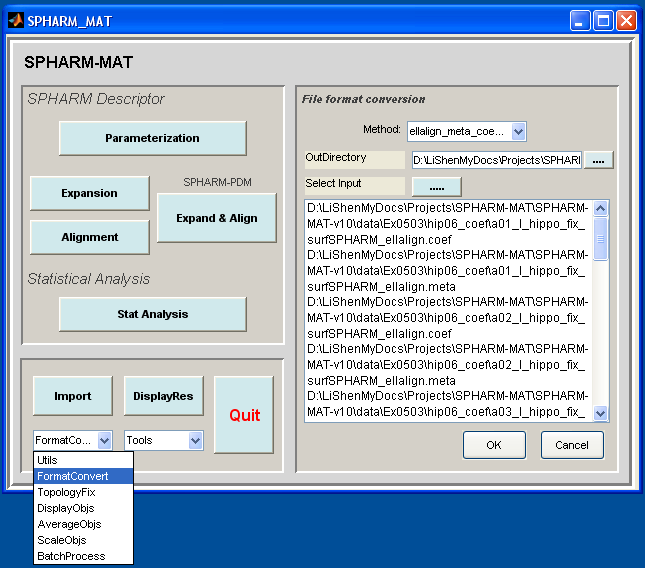
Format Conversion ellalign_meta_coef2reg¶
Task
Convert COEF coefficient format and META surface format generated by SPHARM-PDM to matlab format
Input
SpharmMatDir/data/Ex0503/hip06_coef/*.coef and SpharmMatDir/data/Ex0503/hip06_coef/*.meta
Output
SpharmMatDir/data/Ex0503/hip07_reg/*_des.mat: SPHARM models in original coordinate system
SpharmMatDir/data/Ex0503/hip07_reg/*_reg.mat: SPHARM models in FOE aligned coordinate system
Steps
Make an output directory SpharmMatDir/data/Ex0503/hip07_reg/
Run SPHARM_MAT.m under Matlab
Select FormatConvert under the Utils pop-up menu (bottom-left corner)
Be sure to run this experiments twice by setting different configurations in the right panel as follows
- Run 1: let Method be meta_coef2des
- Run 2: let Method be ellalign_meta_coef2reg
In the right panel, select SpharmMatDir/data/Ex0503/hip07_des/ as OutDirectory
In the right panel, click ..... button next to Select Input, and select all the *.coef *.meta files under SpharmMatDir/data/Ex0503/hip06_coef/ as input files
Click OK button (See Screen shot for Format Conversion ellalign_meta_coef2reg.)
Notes
- This conversion packages *.coef (SPHARM coefficents) and *.meta (Surface reconstruction created by SPHARM-PDM) into a single *.mat file: either (1) *_des.mat for SPHARM models in the original coordinate system, or (2) *_reg.mat for SPHARM models in the FOE aligned coordinate system. Therefore, the surface in the resulting *_des.mat or *_reg.mat file is a SPHARM reconstruction generated by SPHARM-PDM instead of the original object surface. See also Data Structure and Format Conversion for SPHARM Models.
- SPHARM-PDM stores only real parts of the coefficients in *.coef files. See also SPHARM Expansion PDM Difference.
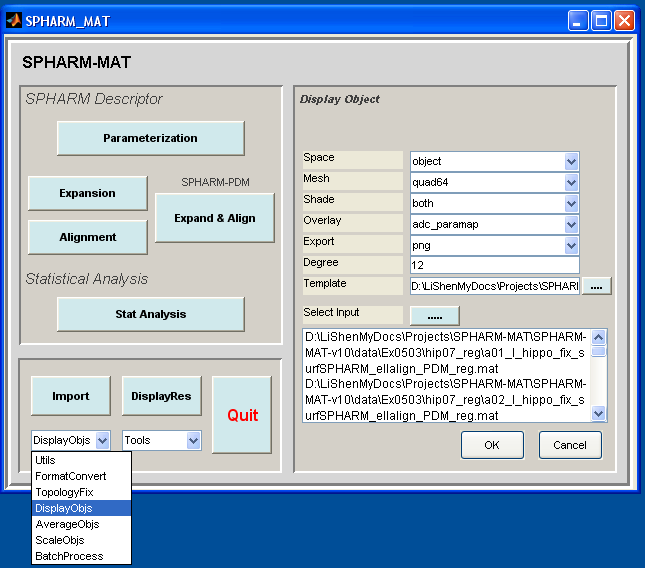
Surface Visualization (quad64)¶
Task
Visualize SPHARM reconstructions before and after FOE alignment
Input
SpharmMatDir/data/Ex0503/hip07_reg/*.mat
Output
SpharmMatDir/data/Ex0503/hip07_reg/PNG/*.png
Steps
Select DisplayObjs under the Utils pop-up menu (bottom-left corner)
Be sure to run this experiments twice by setting different configurations in the right panel as follows
- Run 1 (SPHARM reconstruction using coefficients up to degree 1): let Space be object, Mesh be quad64, Shade be both, Overlay be adc_paramap, Export be png, Degree be 1
- Run 2 (SPHARM reconstruction using coefficients up to degree 12): let Space be object, Mesh be quad64, Shade be both, Overlay be adc_paramap, Export be png, Degree be 12
In the right panel, click ..... button next to Select Input, and select all the SpharmMatDir/data/Ex0503/hip07_reg/*.mat files as the input files
Click OK button (See Screen Capture for Surface Visualization (quad64))
Notes
The visualization results are saved as PNG files under SpharmMatDir/data/Ex0503/hip07_reg/PNG/.
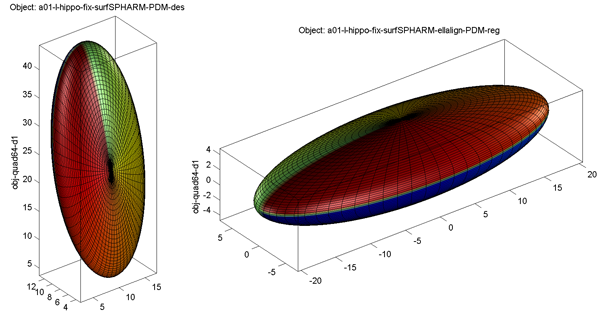
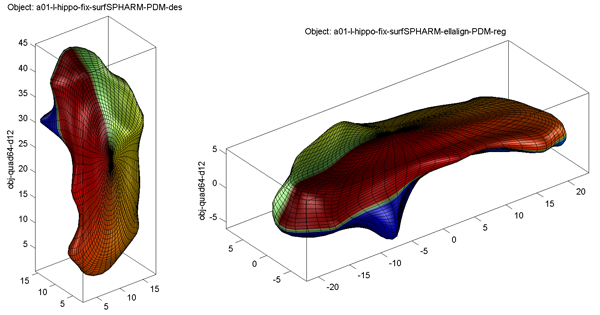
Sample alignment results are shown below: original model on the left and FOE aligned model on the right
- Surface Visualization (quad64, Degree 1): SPHARM reconstructions using coefficients up to degree 1 (i.e., FOE)
- Surface Visualization (quad64, Degree 12): SPHARM reconstructions using coefficients up to degree 12
See DisplayObjs Overlay Option for information about color-coded visualization of the underlying parameterization.
Relevant Information¶
Useful Tips